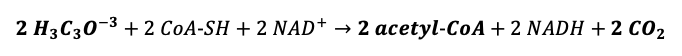Metabolosomes: Difference between revisions
m Fix typo |
|||
| Line 47: | Line 47: | ||
== Scientific Background == | == Scientific Background == | ||
== Scientific Background == | |||
Unlike eukaryotes, prokaryotes do not have membrane-bound organelles. In other words, their organelles do not have membranes. Instead, prokaryotes like bacteria construct compartments out of proteins. A metabolosome is an example of such a compartment, officially referred to as a bacterial microcompartment (BMC). BMCs are composed of a polyhedral protein shell around 100-200 nm large that looks similar to a virus capsid. The proteins that make up the shell typically have (semipermeable) pores that allow different compounds (substrates and products) in and out of the organelle. They concentrate enzymes and proteins involved in specific metabolic processes in one spot, ensuring their success. The protein shell also acts as a physical barrier, allowing BMCs to conduct metabolic reactions that create toxic or unstable intermediate compounds. | |||
Metabolosomes are a catabolic type of BMC, and are involved in the degradation of different carbon sources such as glycerol and amino acids. There is no clear evidence that the process of breaking down glucose takes place in metabolosomes, however it is not unreasonable to assume that it could do so. | |||
==Aerobic cellular respiration (glucose -> ATP)== | |||
The actual process of breaking down glucose for energy takes place in several steps in both prokaryotes and eukaryotes. The only difference is where some of the enzymes are located and organized. Below is a summary of the various reactions with the full names of the molecules and the chemical formulas/abbreviations. | |||
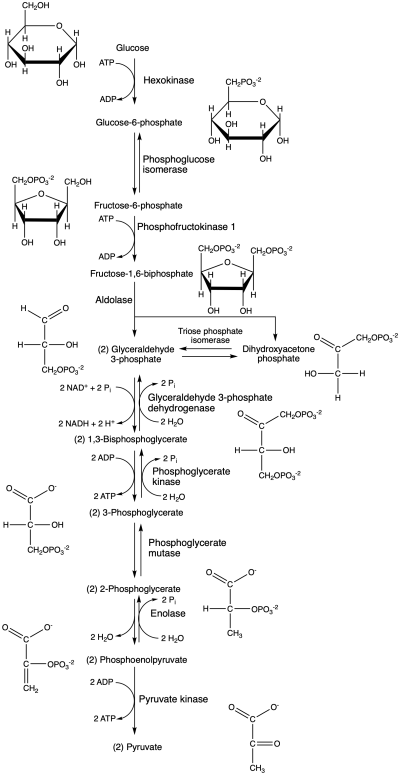
'''Glycolysis [1]:''' | |||
Glycolysis takes place in several steps, catalyzed by 10 different enzymes with 11 intermediate compounds. The end result is one glucose produces two pyruvate molecules, two ATP molecules and two NADH molecules (which are later converted to 4-5 ATP molecules). | |||
The net reaction looks like this: | |||
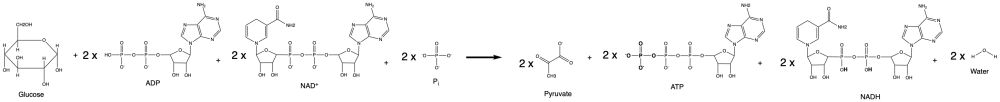
[[File:Glycolysis net reaction formula.png|500px|Figure 1: The carbon molecules are bold and the ATP produced is highlighted in green]] | |||
The net reaction with the full names for the different molecules above: | |||
''Glucose + 2 Adenosine DiPhosphate + 2 Nicotinamide Adenine Dinucleotide + 2 inorganic Phosphate → 2 pyruvate + 2 Adenosin TriPhosphate + 2 Nicotinamide Adenine Dinucleotide Hydrated + 2 water'' | |||
The chemical structures look like this: | |||
[[File:Glycolysis net reaction.png|1000px|The chemical structures for the different compounds in the net reaction for glycolysis]] | |||
If you'd like to learn more about glycolysis, visit https://www.tuscany-diet.net/2018/02/06/glycolysis/. The full glycolysis reaction: | |||
[[File:Glycolysis full.png|400px|The full glycolysis reaction with all intermediates (the chemical structures) and enzymes as well as substituents (ATP, ADP, H2O etc) included.]] | |||
'''Linker reaction [2]:''' | |||
''2 Pyruvate + 2 Coenzyme A + 2 Nicotinamide Adenine Dinucleotide + 2 inorganic Phosphate → 2 pyruvate + 2 Adenosin TriPhosphate + 2 Nicotinamide Adenine Dinucleotide Hydrated + 2 water'' | |||
[[File:Linker reaction.png|The linker reaction between glycolysis and the citric acid cycle/krebs cycle. Converts pyruvate to acetyl-CoA.]] | |||
'' | |||
Rough draft'' | |||
2〖H_3 C〗_3 O^(-3)+2CoA SH+2NAD^+→2 acetyl CoA+2NADH+2CO_2 | |||
Krebs cycle [3]: | |||
2 acetyl CoA+6NAD^++2FAD+2GDP+〖2P〗_i+2ADP+ 4H_2 O→2 CoA SH+ 6NADH+ 2FADH_2+4H^++2GTP+ 4〖CO〗_2 | |||
Electron transport chain and oxidative phosphorylation [4]: | |||
(2+2+6)(NADH+H^++〖1/2 O〗_2 )+2(FADH_2+1/2 O_2 )+2GTP→10×2.5ATP+5O_2+2×1.5ATP+O_2+2ATP | |||
Total ATP produced: | |||
2ATP+25ATP+3ATP+2ATP=32ATP | |||
Although the actual maximum amount of ATP possible varies between species and tissues due to differences in the amount of hydrogens pumped across the mitochondrial membrane by the electron transport chain. In other words, the amount of ATP generated per NADH and FADH2 molecule depends on the species. The total amount is commonly stated as being between 36-38 ATP. | |||
Revision as of 13:27, 22 November 2023
| Organelle Details | |
|---|---|
| Metabolosomes | [[File:|center|100px]] |
| [[File:|300px]] | |
| {{{image_caption}}} | |
| Base Cost (MP) | 45 |
| Requires Nucleus | No |
| Processes | Protein Respiration |
| Enzymes | None |
| Size (Hexes) | 1 |
| Osmoregulation Cost | 1 |
| Storage | 0.5 |
| Unique | No |
| Upgrades | None |
| Internal Name | metabolosome |
Metabolosmoes perform Protein Respiration, a form of Aerobic Respiration inferior to that of Mitochondria. It is the conversion of Glucose into ATP with the use of Oxygen.
Requirements
No requirements
Processes
Protein Respiration: Glucose → ATP @ Oxygen
Protein Respiration is a method of energy production performed by Metabolosomes. An input of glucose is needed for this to take place, and the rate of the process scales with the amount of environmental Oxygen.
Modifications
No modifications.
Effects
No effects.
Upgrades
No upgrades.
Strategy
TBA
Scientific Background
Scientific Background
Unlike eukaryotes, prokaryotes do not have membrane-bound organelles. In other words, their organelles do not have membranes. Instead, prokaryotes like bacteria construct compartments out of proteins. A metabolosome is an example of such a compartment, officially referred to as a bacterial microcompartment (BMC). BMCs are composed of a polyhedral protein shell around 100-200 nm large that looks similar to a virus capsid. The proteins that make up the shell typically have (semipermeable) pores that allow different compounds (substrates and products) in and out of the organelle. They concentrate enzymes and proteins involved in specific metabolic processes in one spot, ensuring their success. The protein shell also acts as a physical barrier, allowing BMCs to conduct metabolic reactions that create toxic or unstable intermediate compounds.
Metabolosomes are a catabolic type of BMC, and are involved in the degradation of different carbon sources such as glycerol and amino acids. There is no clear evidence that the process of breaking down glucose takes place in metabolosomes, however it is not unreasonable to assume that it could do so.
Aerobic cellular respiration (glucose -> ATP)
The actual process of breaking down glucose for energy takes place in several steps in both prokaryotes and eukaryotes. The only difference is where some of the enzymes are located and organized. Below is a summary of the various reactions with the full names of the molecules and the chemical formulas/abbreviations.
Glycolysis [1]:
Glycolysis takes place in several steps, catalyzed by 10 different enzymes with 11 intermediate compounds. The end result is one glucose produces two pyruvate molecules, two ATP molecules and two NADH molecules (which are later converted to 4-5 ATP molecules).
The net reaction looks like this:
![]()
The net reaction with the full names for the different molecules above:
Glucose + 2 Adenosine DiPhosphate + 2 Nicotinamide Adenine Dinucleotide + 2 inorganic Phosphate → 2 pyruvate + 2 Adenosin TriPhosphate + 2 Nicotinamide Adenine Dinucleotide Hydrated + 2 water
The chemical structures look like this:
If you'd like to learn more about glycolysis, visit https://www.tuscany-diet.net/2018/02/06/glycolysis/. The full glycolysis reaction:
Linker reaction [2]:
2 Pyruvate + 2 Coenzyme A + 2 Nicotinamide Adenine Dinucleotide + 2 inorganic Phosphate → 2 pyruvate + 2 Adenosin TriPhosphate + 2 Nicotinamide Adenine Dinucleotide Hydrated + 2 water
Rough draft
2〖H_3 C〗_3 O^(-3)+2CoA SH+2NAD^+→2 acetyl CoA+2NADH+2CO_2
Krebs cycle [3]: 2 acetyl CoA+6NAD^++2FAD+2GDP+〖2P〗_i+2ADP+ 4H_2 O→2 CoA SH+ 6NADH+ 2FADH_2+4H^++2GTP+ 4〖CO〗_2
Electron transport chain and oxidative phosphorylation [4]:
(2+2+6)(NADH+H^++〖1/2 O〗_2 )+2(FADH_2+1/2 O_2 )+2GTP→10×2.5ATP+5O_2+2×1.5ATP+O_2+2ATP
Total ATP produced: 2ATP+25ATP+3ATP+2ATP=32ATP
Although the actual maximum amount of ATP possible varies between species and tissues due to differences in the amount of hydrogens pumped across the mitochondrial membrane by the electron transport chain. In other words, the amount of ATP generated per NADH and FADH2 molecule depends on the species. The total amount is commonly stated as being between 36-38 ATP.